Systems Biology: Using Computers to Kill Cancer Cells
Cell death is a biological process that we need to maintain balance in the body. It helps us to develop organs and limbs but if it gets messed up, different types of disease can occur. Too much cell death can cause neurodegenerative diseases like Alzheimer’s whereas too little can lead to cancer.
There are two ways that cell death can be activated. 1. By an external signal – this is termed the "extrinsic pathway" or 2. by a signal from within the cell itself – this is termed the "intrinsic pathway".
Both cell death pathways are driven by proteins called Caspases. Tight regulation of these proteins is important for effective cell death. There are two different types of caspases, classified by their role in the cell death pathway. "Initiator" caspases are activated when the cell receives a signal to die. Once activated, initiator caspases go on to activate the "executioner" caspases which regulate the breaking down of the cell.

This work focuses on the intrinsic cell death pathway and what regulates it, and aims to figure out a way to take advantage of the intrinsic cell death pathway in a disease setting – for example, to activate more effective cell death in cancer. There are a number of different proteins which regulate the intrinsic cell death pathway that are outlined in Figure 1.

I created a computational model of the intrinsic cell death pathway. The input of the model is a DNA damage agent (like a cancer therapy) that activates BAX and the read out of the model is activation of the executioner caspase (which we take to mean cell death). We aim to figure out how BAX and SMAC contribute to the activation of cell death and how we could take advantage of these proteins to create effective therapies.
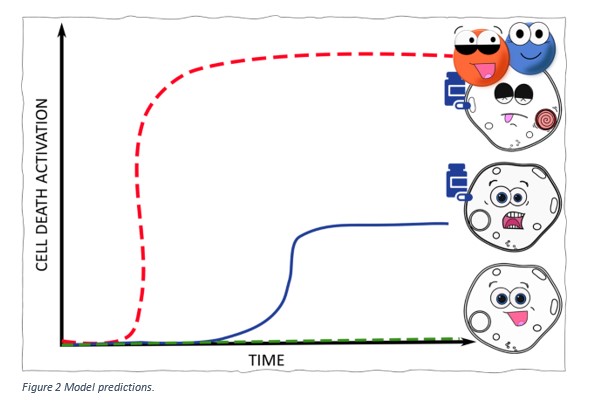
Our model simulations predict that cells which have a higher BAX or SMAC level compared to those cells that don’t will:
- Have activation of intrinsic cell death with a lower concentration of drug
- Have more cell death activated.
Figure 2 shows these predictions. The green line represents cells that are not treated with the drug and show no activation of cell death. The blue line shows that cells which have low levels of BAX/SMAC and when treated with the drug only show low levels of cell death. The red line represents cells that have high expression of BAX/SMAC. When these cells are treated with the drug, our model predicts that higher levels of cell death will be observed at a lower concentration of drug, compared to the green and blue cells.
The method of systems biology, which I use throughout my model construction, relies on several rounds of model predictions and experimental investigation and validation of those predictions. So, I carried out several experiments to validate the model predictions that cells with higher levels of BAX/SMAC show a higher level of intrinsic cell death activation.

So why are models like mine important for cancer patients? Figure 3 shows an example of patient specific simulations. To carry out these simulations, specific information can be gathered from each patient’s tumour- for example, BAX/SMAC concentration. The patient’s information is then used as input for the model. Patient simulations help us to identify which patients could respond to which therapy. According to this example: Patient 3 expresses a high level of BAX and would respond better a drug which activates cell death. Patient 1 and 2 might benefit from a combination of therapies including an agent that increases BAX levels to match those of patient 3, enhancing the activation of cell death in their tumours.

I had great fun presenting a lay poster of this work at the Choirs for Cancer event on World Cancer Day 2020.
The manuscript under review is entitled ‘BAX and SMAC Regulate Bistable Properties of the Apoptotic Caspase System’. Check out the preprint version of our paper here:
https://www.biorxiv.org/content/10.1101/867663v1

