‘Spare tire genes’ explain why some genes can be lost by cancer cells
Surprisingly, the majority of human genes can be mutated or deleted in cancer cells without killing the cells. This observation raises a number of questions: which genes can be lost in cancer cells, and why? In a recent study we addressed these questions by analysing data on which genes are required for the survival of over 500 different cancer cell lines. Cancer cell lines are cells taken from patient tumours and grown in the laboratory.
Recent technical advances, such as the development of CRISPR-Cas9 based approaches, have made it possible to observe the consequences of knocking out each gene in a large number of cancer cell lines. Specifically, we can measure how much the loss of each gene affects the growth of each cancer cell line. If loss of a certain gene results in strongly reduced growth of one cell line, that gene can be considered essential for that cell line.
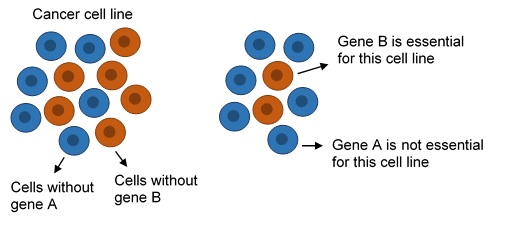
From the DepMap project we obtained growth measurements from 558 different cancer cell lines, which originate from more than 20 different cancer types, and used these to call each of ~16,000 genes either essential (i.e. required for growth) or not essential in each cell line. Notably a gene can be essential in some cell lines and not others. We then calculated the percentage of cancer cell lines in which a given gene was essential.
We found that paralog genes are, in general, essential in fewer cancer cell lines than genes that are not paralogs (these are called singletons). Paralogs are genes that have been duplicated at some point in evolutionary history, resulting in our genome having two copies of (roughly) the same gene—a paralog pair. These paralog pairs, which make up over half the genes in our genome, are a potential source of redundancy, similar to a car having a spare tire. Accordingly, one explanation for why paralogs are less often essential is that their loss, which could be likened to a flat tire for the cell, can be mitigated by a ‘spare tire gene’.

However, we expected that while the loss of one gene from a paralog pair could be tolerated, due to the existence of a ‘spare tire gene’, the loss of both members would not be tolerated - i.e. the car would cease to function if it lost both a tire and its spare. To test this we investigated 1,819 paralog pairs across all the different cancer cell lines. We’ll refer to the two genes that form a paralog pair as A1 and A2, where A2 could be considered the ‘spare tire gene’ for A1.

In all the paralog pairs that we investigated, the A1 gene was essential in some but not all cell lines. This allowed us to compare the status of the A2 gene in cell lines where A1 is and is not essential. Specifically, we compared the expression of A2, which provides a relative measure of the amount of A2 that is present in the different cell lines. For ~13% of the paralog pairs, we found that the expression of A2 was, on average, significantly lower in the cell lines where A1 is essential compared to the cell lines where A1 is not essential. Thus the amount of A2 that is present can explain why loss of A1 is tolerated by some cell lines and not others. This is indicative of a phenomenon called synthetic lethality - the loss of A1 alone can be tolerated, but the simultaneous loss of A1 and A2 is not tolerated.
(Hover mouse over the graph above for more detail.)
Synthetic lethality represents a promising approach for targeted therapies in cancer, in part because gene loss is common in cancer cells, but rare in healthy cells. If we know a cancer cell has lost one member of a synthetic lethal pair, e.g. a ‘spare tire gene’, we can anticipate that it will be particularly vulnerable to loss of the other member. In this study we show that ~13% of paralogs exhibit such a synthetic lethal relationship, and that this explains why loss of some genes is sometimes, but not always, tolerated by cancer cells.
Although the focus of our work was not on the identification of new targets for cancer therapies, our findings suggest that certain types of paralogs are more likely to be involved in synthetic lethal relationships than others. This may ultimately help guide the search for new therapeutic targets.
Research team:
This study was carried out by Barbara De Kegel, a PhD student in SBI, and Colm J. Ryan, an assistant professor in the UCD School of Computer Science.
You can read the paper here: https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1008466
About the Author:
Barbara De Kegel is a computational cancer biology PhD student in Dr. Colm Ryan’s group at SBI & the UCD School of Computer Science. Her background is in computer science and she joined SBI after working as a software engineer for Havok (Microsoft), who develop physics components for video games.

